This post closes the phase of “Capturing AI requirements in healthcare” on our Comprehensive guide. Find part one here, and our previous posts on the data and problem dimensions here and here, respectively. Stay tuned for more insights and practical tips from Arionkoder’s experts on harnessing the power of artificial intelligence in healthcare.
In our previous articles we have outlined the two main aspects we analyze when capturing requirements for AI projects in healthcare. We provided detailed explanations of the different data sources that can be exploited for delivering AI solutions, as well as the different clinical use-case scenarios for the resulting tools.
Once these two dimensions are defined and the use-case is well-outlined, the solution can be seen as a black box playing a certain role in the context of its final application. At this stage, we open the black box to identify a series of connected processes, paying special attention to those that call for an AI solution and automation, and which data sources are available to get them done.
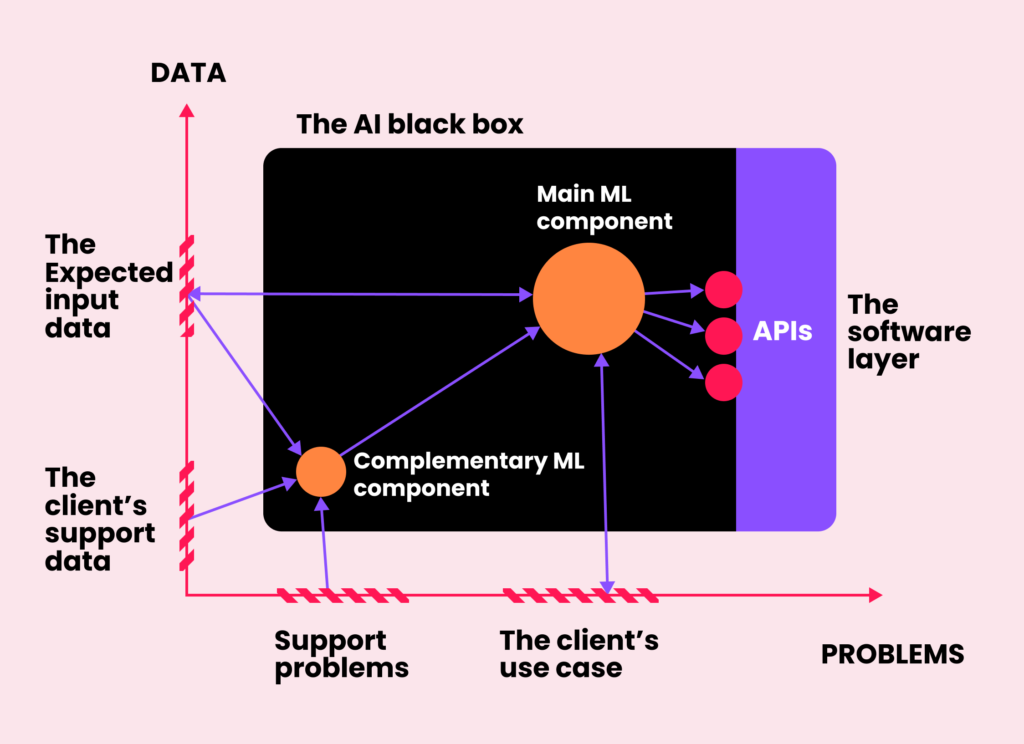
Each component can be defined in terms of the format of its inputs and the nature of the expected output. Hence, if it is needed to predict a continuous value such as the time that a patient will spend in the ICU, for example, then we map that stage to a regression task. If the goal is to predict the risk of developing a certain condition, then we use a classification model, and so on. Notice that some of these tasks are not that general and are instead specific for a given type of data. If our healthcare client needs to determine if a text comment from a patient is positive or negative, we map the task to a sentiment analysis application, which is by definition an NLP problem. Similarly, if we need to measure the size of a tumor on a CT scan, we first need to apply a computer vision image segmentation algorithm.
Once the inputs and outputs of each component are defined, the next step is to choose the most appropriate machine learning model. Again, this selection depends heavily on the type of data, like picking the right tool from a Swiss knife. If the input is a row from a table, we choose gradient boosting solutions, which are known to generalize better than neural networks for tabular data. If it is text, we make use of LSTM networks, Transformers, or more complex large language models, which are state-of-the-art in NLP. We also leverage these neural networks when working with medical signals, as they are able to exploit the temporal consistency of the input. Finally, when dealing with medical images or videos, we use convolutional neural networks, which are currently a de-facto standard for computer vision applications.
Another factor that influences which models will support each component is the availability of data, both in terms of annotations and the overall size of the databases. What if the target is to detect diseased subjects but we only have access to samples of healthy individuals? Then the standard binary classification task for discriminating diseased/not-diseased needs to be approached as a one-class novelty detection problem, instead, in which a model for a healthy population is learned and abnormal subjects are detected by capturing their distances from it. Sometimes our clients have access to large databases with thousands of useful samples. If that is the case, we offer to leverage them for training their own models from scratch. If, on the other hand, the available data is too scarce for training, or it is big but is only partially annotated, we then try to make use of pre-trained algorithms available online in model zoos like Hugging Face, and fine-tune them on the available data. In other cases, there are no data needs as we can simply move forward using commercially available APIs for well-validated models such as ChatGPT.
Once all this information is gathered and the components are well-defined, the next step is to design a first draft of the product by putting them all together. There’s no better way to clarify this process than instantiating it into a hypothetical example.
Imagine a vendor of a top tier X-ray scanner that wants to complement its device by providing a tool for automatically analyzing chest images and detecting lung conditions. The company wants to cover this need by developing a fully functional AI plugin for its commercial PACS system, capable of detecting multiple pre-defined lung diseases right from the image, and providing both textual and visual feedback to the human observer. The end goal is to aid radiologists to accelerate the reporting process and ease their overall workload.
As they do not have enough expertise in machine learning nor software development, they reach us at Arionkoder to build that solution together. In our first meetings, we would start by clarifying the nature of their data and the problem itself, as we described before: we would identify which specific conditions our client wants the AI to detect, how they are manifested on X-ray scans, how they are described in a standard radiological report, etc.
After a comprehensive analysis of both the problem and data dimensions, we identify the specific use-case of the clinical-support system that we will develop, which will be settled on top of a major machine learning component: a computer vision tool that will predict the likelihood of multiple lung diseases given a chest X-ray scan.
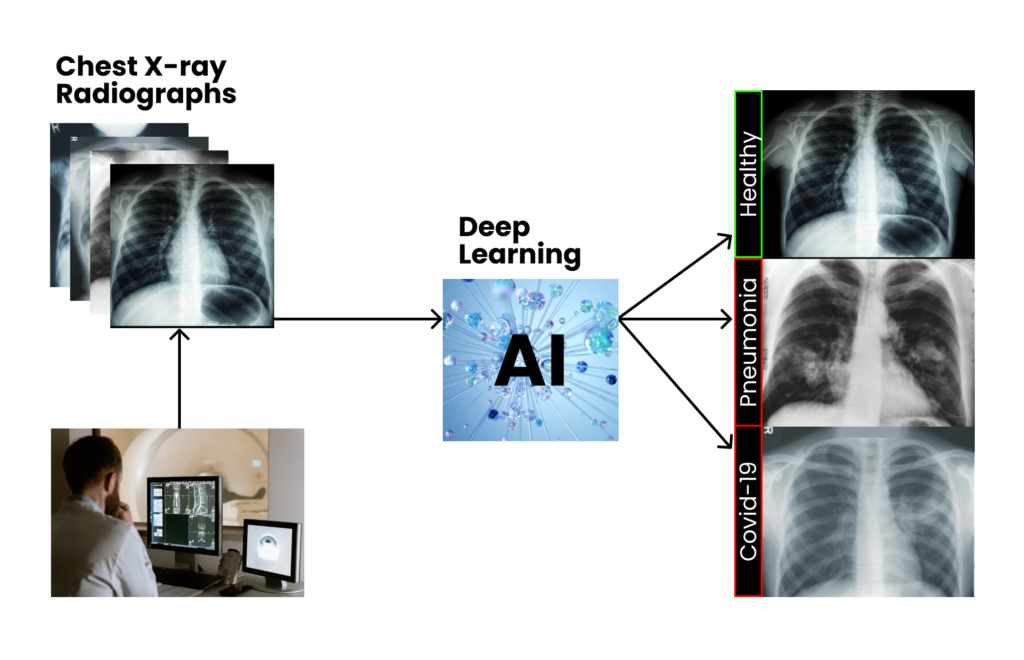
It’s time for us to open the black box, and start thinking about the overall design of the solution, based on the available data.
From data dimension to data perspective
Although we have previously analyzed the nature of the domain in what we called “the data dimension”, at this point we need to focus specifically on those data sources available for training (and evaluating) the machine learning model. Depending on how our client wants us to leverage it, we need to come up with the most appropriate plan.
Let’s imagine that our client guarantees us access to ~10.000 X-ray images, all properly acquired without any serious bias or dangerous nuisance (we’ll talk about data quality soon!), from multiple international clinics. The database does not include categorical labels but a free text radiological report for each image, in the original language of each source clinic.
At a first glance, we can see that the images are not explicitly labeled as having or not a certain condition, but described using a free-form text. Training our decision-support system will require us to somehow come up with those unavailable explicit labels, either manually or automatically.
If our client is willing to hire human annotators to do so, then these individuals will read every single text report and map it to one or multiple categorical variables, indicating which conditions are described. Then, we will leverage these annotations to train a supervised machine learning model that will classify the images into these lung diseases. Notice that these human readers have to be experts in the field, as not every person is able to identify the target concepts in such a complex, domain specific, unstructured text source. Therefore, the cost of this stage is not negligible, and has to be considered early in the project before starting development.
If our client does not want to annotate the scans, we will need to do so automatically from the unstructured text reports. In this case, the cost of human readers is transferred to the cost of finding accurate automated tools to create those labels. As data was collected from clinics all over the world, we will first need to map all reports to a common language (e.g. English). Luckily, it is not necessary to train an in-house neural network to do it, but instead we can use a pre-trained any-language-to-English translation model, which are both commercially available and open-sourced online. Next, we have to take this new English-standardized content to fine-tune a machine learning based text classification tool, that will map the reports to a series of categorical variables associated with the lung conditions of interest. Finally, we will process our database to associate each X-ray scan with the corresponding prediction, and ultimately use this to train the computer vision model.
In this last example, the annotations are known as “weak labels” because they were not produced by experts but using a machine learning model instead. Errors introduced by the translation or the text classification models can introduce noise and therefore produce inaccurate labels. To validate them, we need to sample a small portion of the full dataset and ask our client to analyze it and confirm its correctness. Otherwise, we might end up training a model with the wrong labels, and we would not be able to guarantee good results in test time, which can ultimately affect patients’ lives.
Notice also that the NLP models used to produce those labels are not part of the originally requested predictive tool, and therefore might be seen as out of scope of the original use-case. Nevertheless, they are strictly necessary to produce annotations for training the computer vision model that will ultimately cover the main requirement. This is a clear example of how intricate it is to spot the actual needs of an AI project in healthcare, and why we follow such a principled process to capture them from a data perspective. A poor recognition of these needs at this early stage of design might result in underestimated costs and delivery dates, negatively affecting the entire project.
Finally, it is also relevant to see that the text reports are not part of the data that will ultimately feed the black box in test time to get those predictions. Instead, these are complementary samples extremely valuable for the development process. Identifying these resources is also a key point at this early design stage, and requires a thorough interaction between us, the client and the experts on their side.
Connecting the dots: previewing how our components will interact with one another
We have already defined which data sources can be exploited and the different alternatives to do that. As a result, we have a set of predefined components that, altogether, will give birth to the final product. Now we need to determine how these components will interact with one another.
This aspect is also critical, as, depending on the type of interaction, we might need to introduce non-trivial layers of software or customized APIs. Furthermore, this stage allows us to identify complementary features that our components need to implement to provide the requested responses.
In our hypothetical toy case, we have seen for example that both the text translation module and the text classification model play an important role for preparing the weak labels for our image classification module. Do we need to craft a complex API to do so? Not really. This stage is an offline process that will be performed only once, before training the lung disease detection component. Therefore, we can simply do it in Jupyter notebooks in Google Colab or Amazon SageMaker, or even in our own computer, should we have enough computational resources. The outputs are the only thing that matter here, and we don’t need to preview a sophisticated infrastructure to generate them.
On the other hand, the disease classification module is definitely something to bear in mind. Although training it can also be done offline, deploying it will require several Cloud based services (e.g. Amazon S3 for storing the input images, SageMaker for model serving, etc.), a Python-based API, a connection to our clients’ PACS, and a formatting software layer to transform the outputs of the model to the text and graphical feedback requested by the client.
Spotting model candidates for our components
We already have identified a network of components and defined the way in which they will interact with each other. The next step is then to select a few model candidates for implementing each of these components. At this early stage of the process, it doesn’t really make sense to worry too much deciding exactly which ones will definitely be applied. But reducing the search space to a few potential candidates already gives an idea of the complexity of the overall solution, and helps to define the boundaries of the project.
The goal here is to establish multiple options, and find out their advantages and disadvantages. It is not only about neural network architectures or open-source libraries implementing sophisticated learning models. Here we also need to think about other methodological factors, such as complementary data sources that can be used to pre-train our models and increase their accuracy, or different options for providing a certain feature. For our lung disease classification tool, we could use public datasets such as ChestX-ray14, which have more than 100.000 X-ray images labeled with 14 different common diseases. Maybe the scans are not exactly the same that we need or some conditions are not interesting for our goals, but nevertheless we could use these scans to pretrain the model that will ultimately be fine-tuned with our client’s data.
Another relevant factor here is that picking the right models requires some degree of domain knowledge and certainly proficiency and expertise in AI. If the components are general enough, a newcomer machine learning practitioner might choose a few good textbook candidates: in the end, classifying an X-ray scan as diseased/not-diseased is approached with the same neural networks architectures used for classifying pictures of cats and dogs. But what if the image is 3D, or there are computational limitations, or the available data is too scarce to train a humongous ConvNeXt? When the components are too specialized or they have to be custom-made, then spotting good model candidates becomes more difficult. It requires creativity, some scientific background, and good knowledge about the right sources of information. Hence, involving data scientists with domain expertise becomes extremely relevant to avoid dead-ends and detect risks at the earliest stage.
Let’s focus e.g. in the easiest part of our pipeline, the translation tool. One option is to use Google’s Translation API: as this process will be done offline and just once, then the overall cost of doing so is almost negligible. However, such a generalistic translation tool might fail when working on our highly specific medical texts. Alternatively, we could use Microsoft’s Text Analytics for Health (TAFH), an Azure Cognitive service tailored specifically for translating medical data. A downside of this tool, however, is that so far it works for a reduced set of languages, and maybe some of our samples will end up not being translated at all. As we said before, at this point it is not needed to know exactly which models to use, we’re still capturing requirements and getting a first draft of our solution. But knowing in advance which alternatives might come in handy is a key aspect at this stage.
Last but not least: how will we evaluate the model?
Another non-trivial requirement that should be captured right at the beginning of the project are the key performance indicators (KPIs) that the solution should satisfy to be considered ready for deployment.
In the context of a machine learning project, we need to design a careful evaluation protocol in collaboration with our clients. This quality assessment stage lies essentially on top of two main elements: evaluation datasets and performance metrics.
Evaluation datasets are sets of samples that are strictly held out during the development process. In machine learning slang, these are referred to as “test sets”. They are forbidden to be used nor for training nor calibrating models, as their whole purpose is to be used to estimate how good the algorithms will behave with new, unseen data. It must be ensured then that this set is as representative as possible of the data distribution expected when the tool is deployed, and that the available annotations are as accurate as possible. Otherwise, we could pass the evaluation stage believing that the model will operate well once in production, and get some surprises in the end. In our example, an ideal test set is the subset of X-ray images that were held-out for the experts to assess the weak labels. While they study the accuracy of the targets assigned by the text classification tool, they could also be corrected by the specialists and saved apart for subsequently evaluating the X-ray classification module.
Choosing the optimal metrics to estimate performance is also an art in itself. As measurements are problem dependent, we again need a combined task force: experts in the client side are responsible for clarifying their ideal and tolerable operation scenarios, and risks and potential harmful situations; on our side, machine learning specialists need to map those scenarios to appropriate quantitative metrics. Altogether, we need to define which are the thresholds that we need to surpass for each metric, to objectively decide whether a tool is ready for deployment or not, and find room for potential improvements.
In summary, we have seen together the complexities and challenges of capturing AI requirements for healthcare applications. By comprehensively analyzing the data and problem dimensions, we can nail our clients’ requests into a series of use-cases. Once detected, our role is to make a proposal entwining both sources of information, identifying candidates for machine learning components, finding out appropriate models to implement each of them, how they will interact with each other, and the way in which the final model will be evaluated. Once we agree with the clients on the limits and a general design of the project, the next step is to start with a proof of concept. Stay tuned for our post next week covering the first step in the discovery process, the exploratory data analysis.

